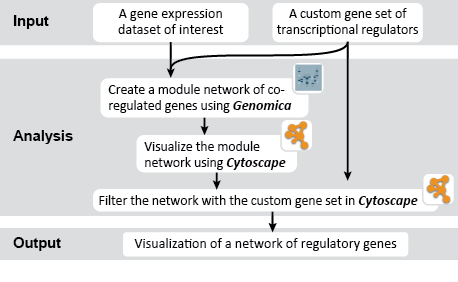
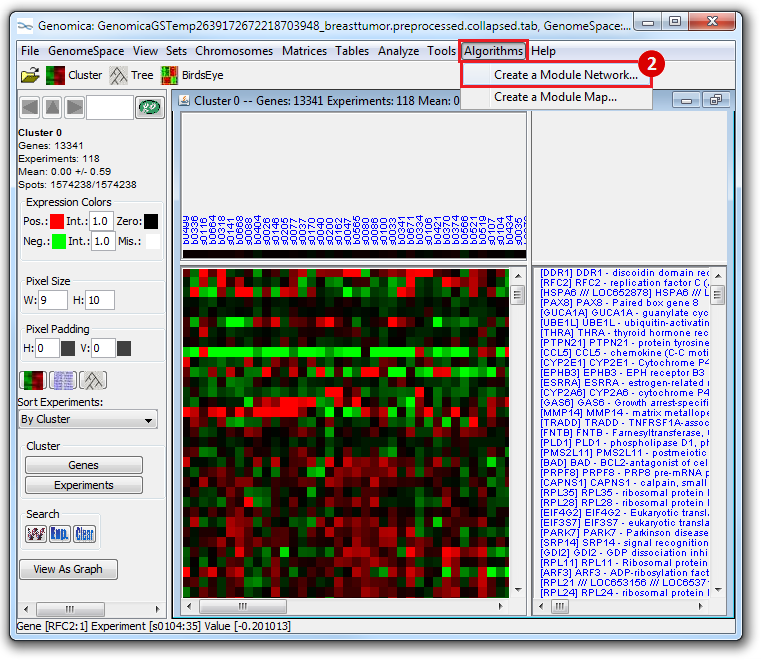
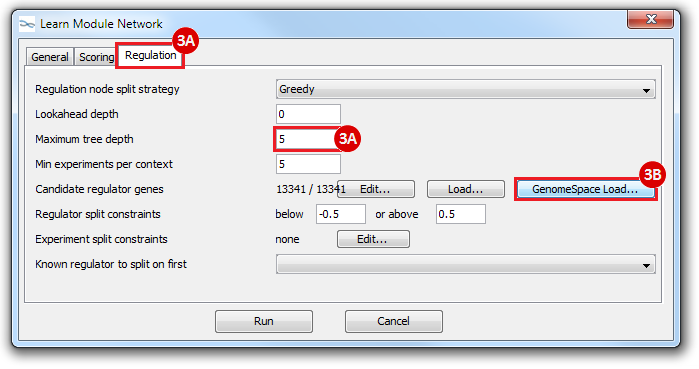
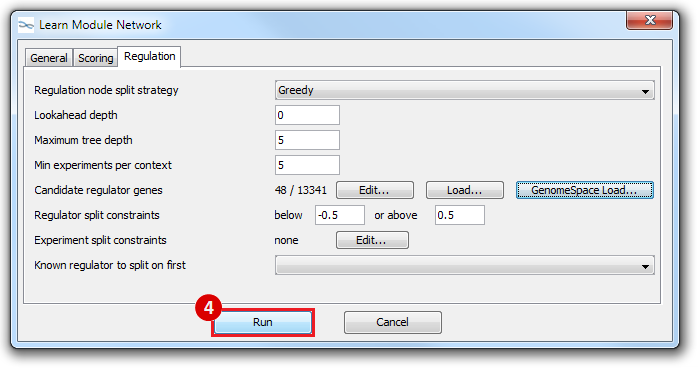
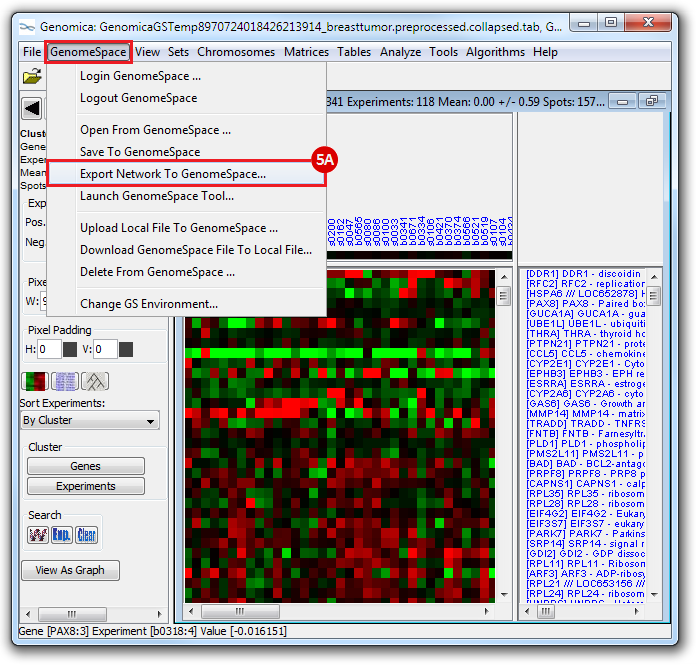
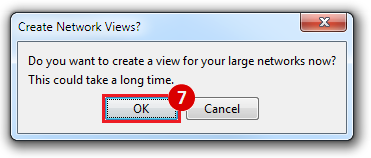
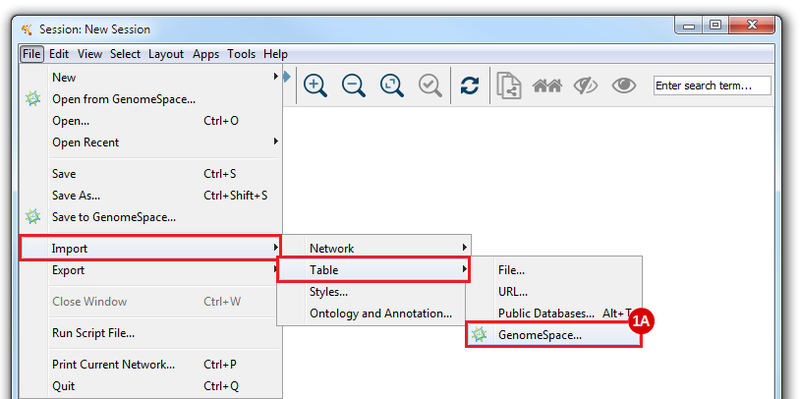
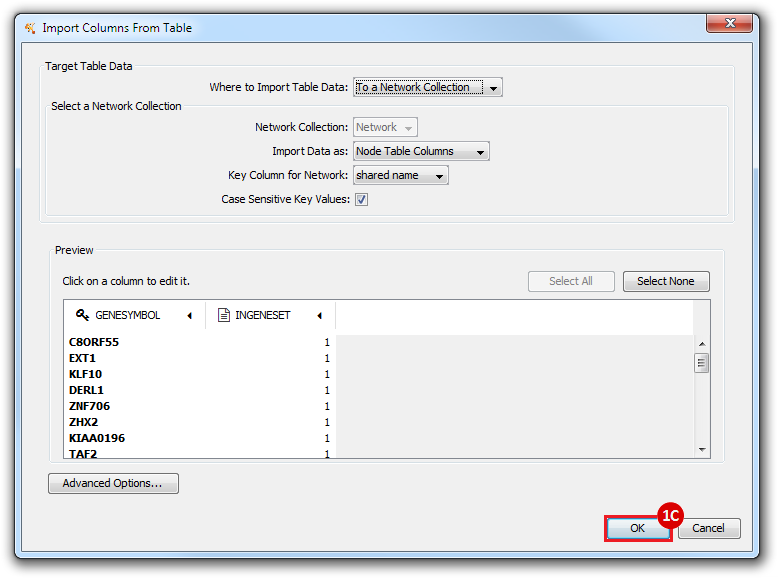
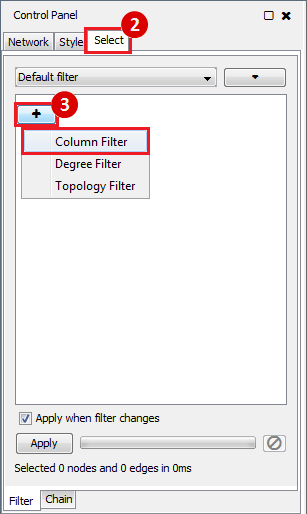
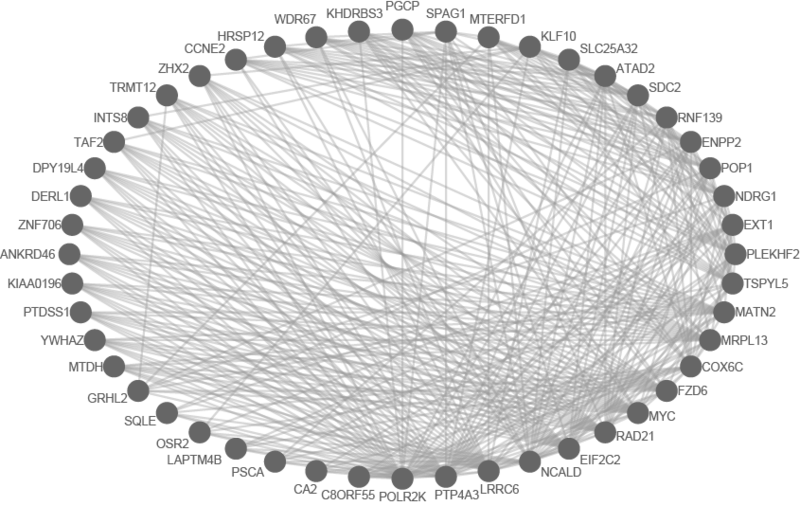
This is an example interpretation of the results from this recipe. First, we created a module network of genes that regulate the stemness signature, by projecting it onto a breast cancer tumor gene expression dataset using Genomica. We then used Cytoscape to visualize the connections between these genes, and to filter a large network down into a subnetwork of just stemness regulators.
We can see from the resulting subnetwork (see below) that, e.g. genes such as POLR2K and MYC are stemness regulators with many connections to other stemness regulators. In contrast, genes such as SQLE or PSCA have fewer connections to other stemness regulators. We can describe POLR2K and MYC as having higher 'degree', than genes such as SQLE or PSCA. These connections in the network imply that perturbing the regulation of POLR2K or MYC would have a larger effect on the embryonic stem cell expression signature (because their multiple connections allow perturbations to propagate in the network) than perturbing other genes that have fewer connections.
 GenomeSpace.org
GenomeSpace.org