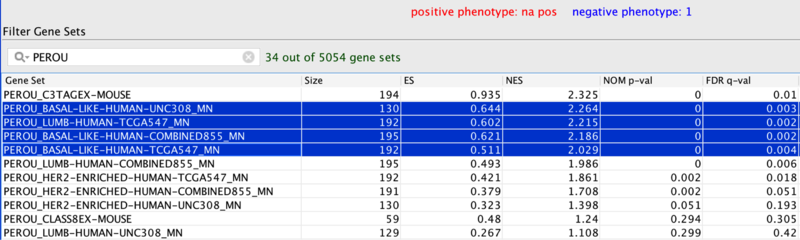
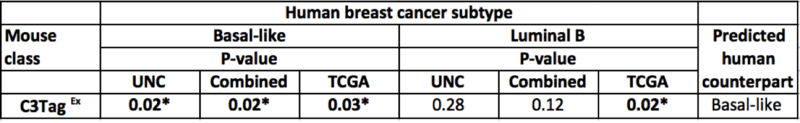
In the original publication (Table 2), the murine C3TagEx class is strongly associated with the human Basal-like subtype(on UNC, TCGA, and combined dataset), and slightly associated with TCGA Luminal B subtype. The present analysis faithfully recapitulates this finding, suggesting that the two mouse models C3 Tag and Wap Tag resemble human Basal-like subtype at the gene-expression level.
Part of Table 2: Gene set analysis of murine classes and human subtypes
Acknowledgement We thank Professor Charles M Perou for providing the data matrices used in the original publication. If you need to use these data for your own research, please cite the original publication below:
Pfefferle AD, Herschkowitz JI, Usary J, Harrell JC, Spike BT, Adams JR et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol 2013;14:R125
 GenomeSpace.org
GenomeSpace.org