This is an example interpretation of the results from this recipe. First, we identify genes which become significantly up-regulated during mild hyperthermia, using GenePattern, resulting in a short list of genes. Next, we were interested in knowing what, if any, functional annotation these genes had - are there specific gene functions which become up-regulate in mild hyperthermia? Are the genes in this condition connected functionally?
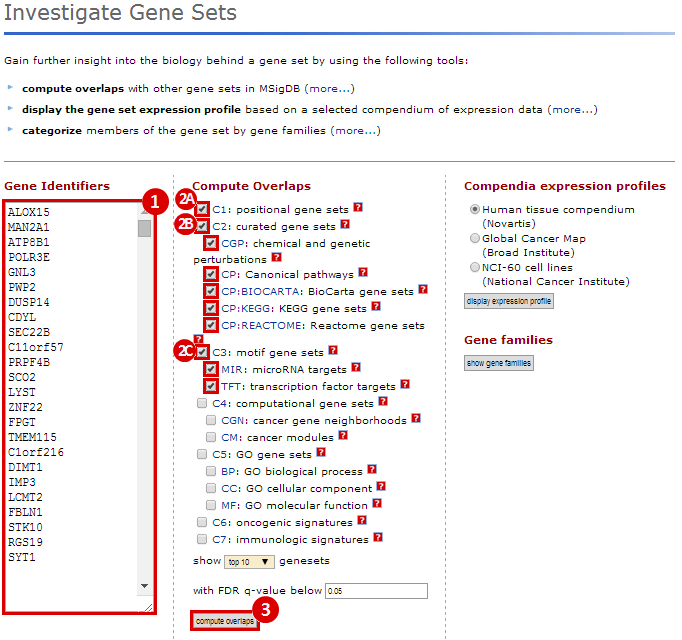
We used MSigDB to probe our dataset for functional annotation. In this case, we used only three collections: C1, C2 and C3. In this example we are most interested in knowing whether our genes are related to chromosomal deletions or amplifications (C1: positional gene set), whether our genes have functions that are reviewed in the literature (C2: curated gene set), and whether our genes share any cis-regulatory motifs (C3: motif gene set).
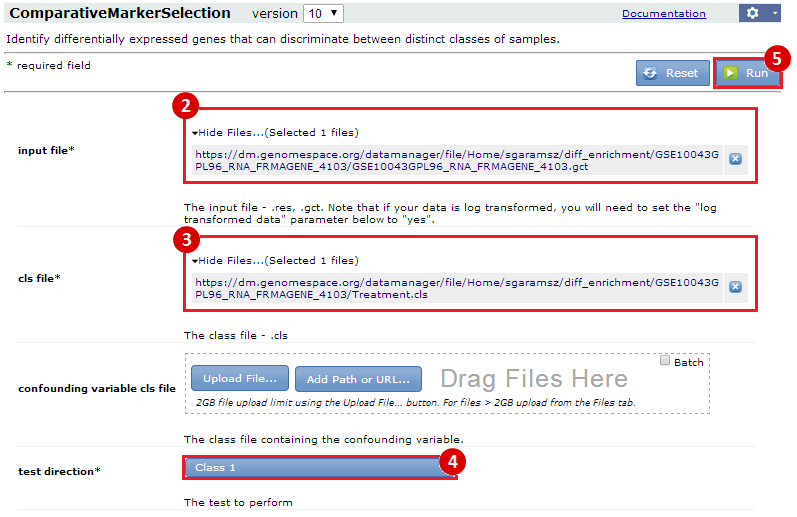
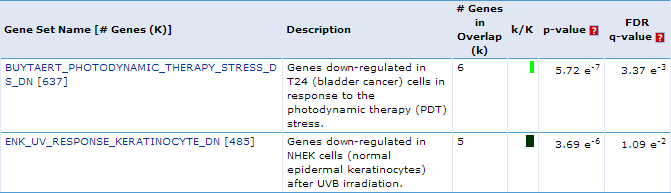
Our first result lists the gene set name and description, the number of our genes which overlap with the gene set, and measures of significance (p-values and q-values). For example, we see that 6 genes out of the 24 we submitted to MSigDB fall into the "BUYTAERT_PHOTODYNAMIC_THERAPY_STRESS_DS_DN" category, which has 637 genes total. This enrichment has a p-value = 5.75e-7. This suggests that genes which become up-regulated during mild hyperthermia are also down-regulated in bladder cancer cells in response to photodynamic therapy stress. This is just one example of a possible interpretation of these results.
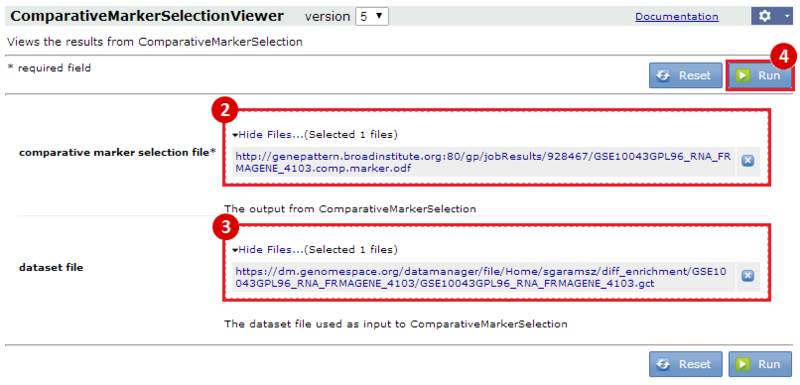
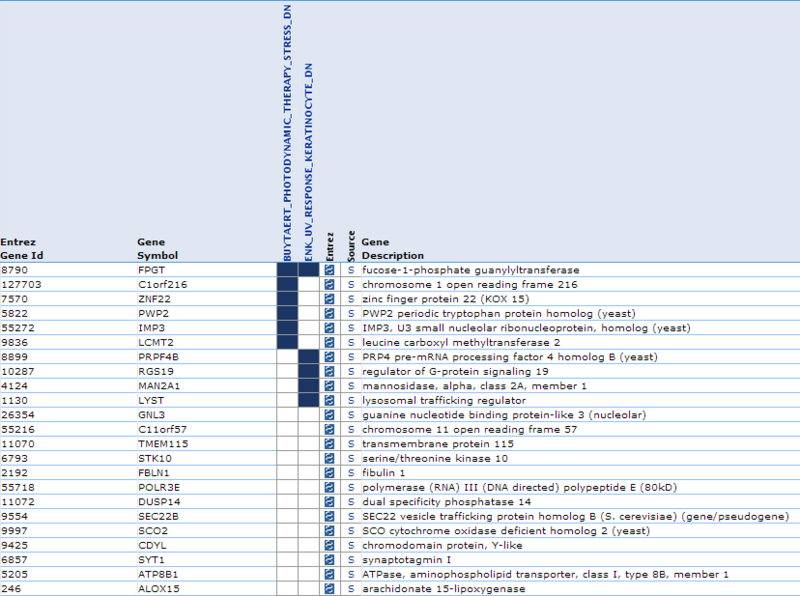
Our second result lists each gene by ID and Symbol, then highlights which of the top categories it is in. For example, lysosomal trafficking regulator (LYST) overlaps with 1 category: ENK_UV_RESPONSE_KERTINOCYTE_DN. If we examine this category, we find that LYST becomes down-regulated in keratinocytes following UVB irradiation.
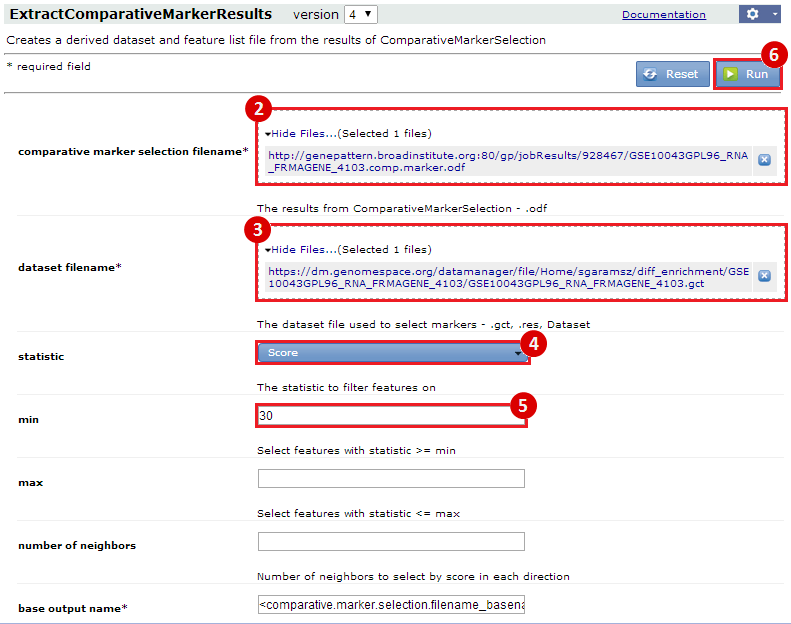
These results suggest that our gene list is enriched for specific functions, which may be associated with the mild hyperthermia condition. However, the results in this example are not necessarily significant and are only a simple representation of possible results.
 GenomeSpace.org
GenomeSpace.org