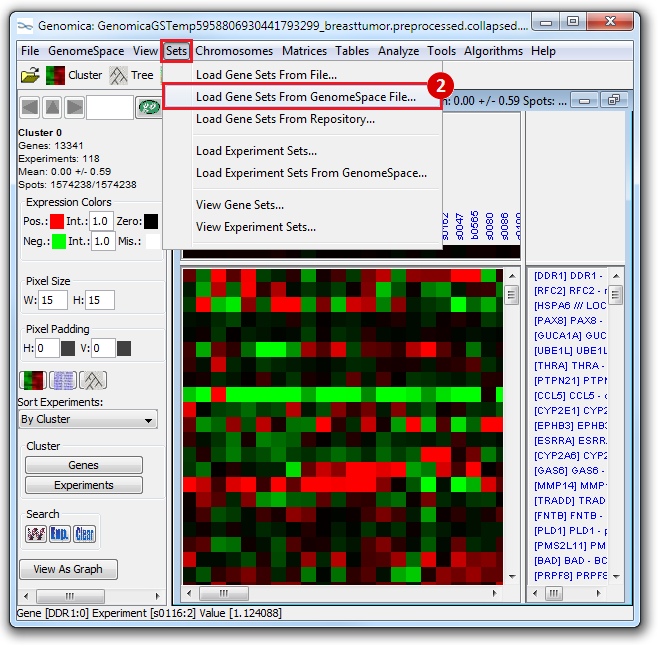
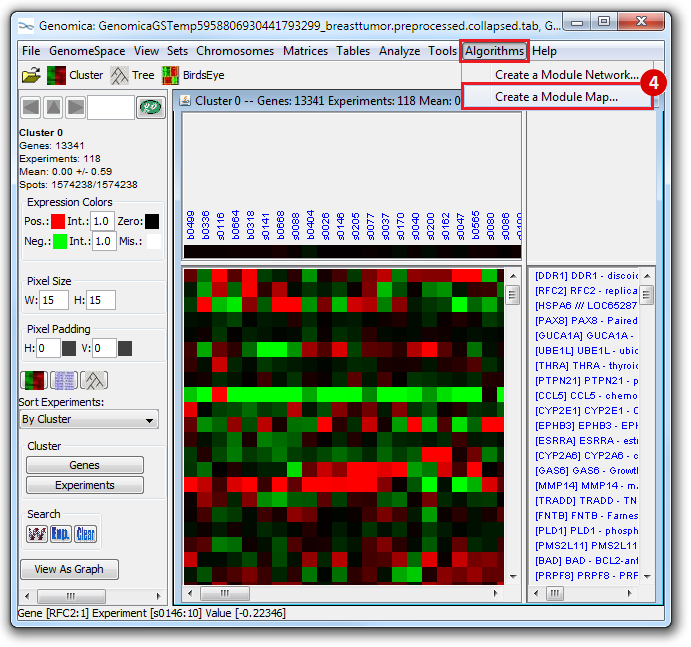
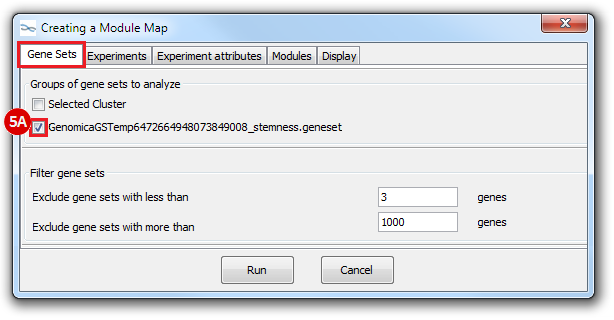
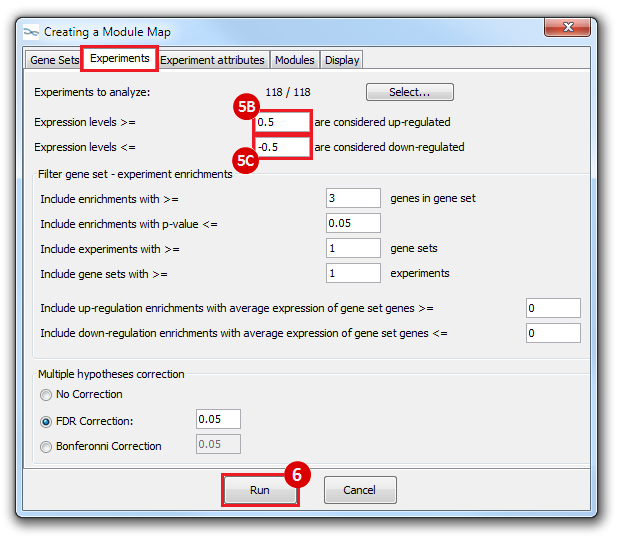
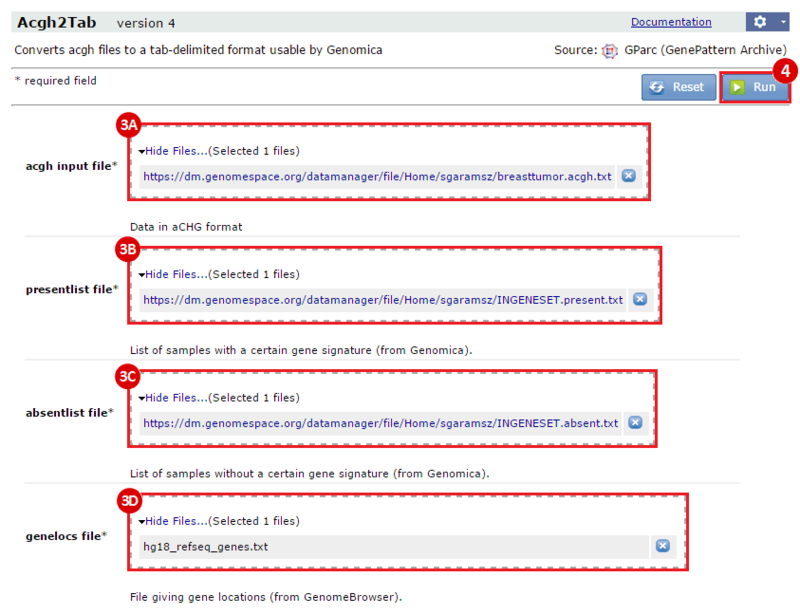
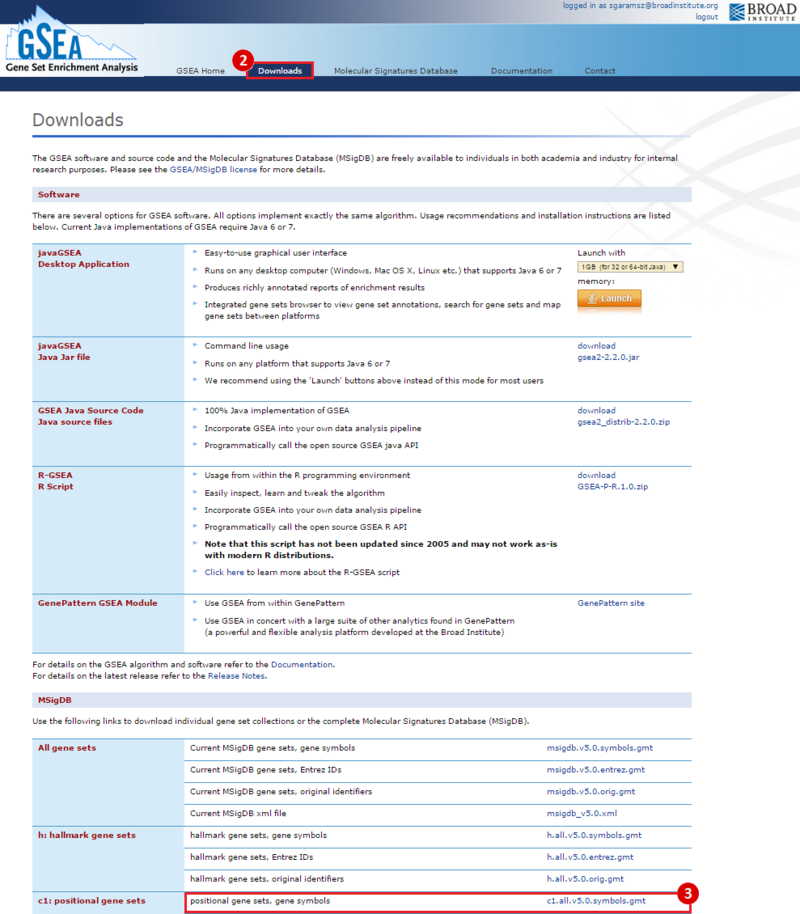
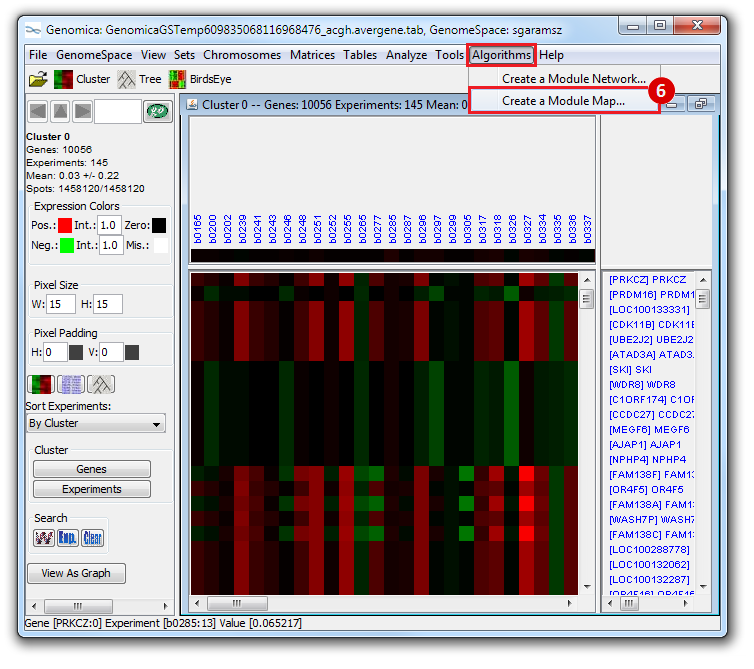
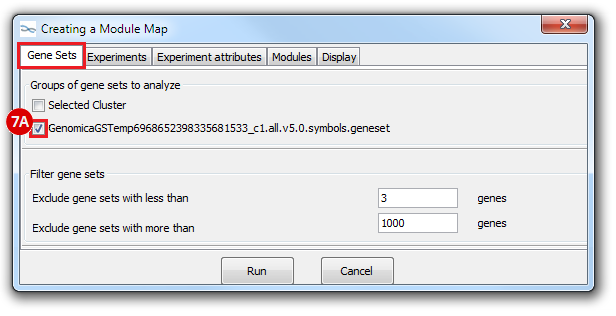
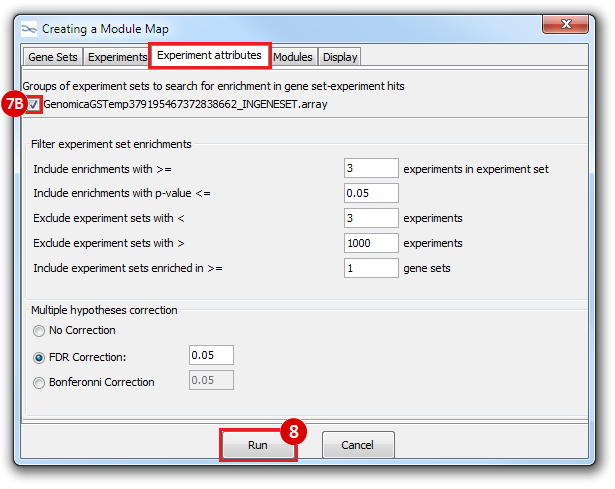
This is an example interpretation of the results from this recipe. First, we identified which breast cancer tumor samples exhibit the stemness signature and which did not, and used this classification to normalize copy number variation data. Next, we identified transcriptional regulators of the copy number variations by overlapping the copy number dataset with a gene set collection from MSigDB. Finally, we identified regulators of the stemness signature by identified genes with concordant profiles in the breast cancer tumor sample gene expression dataset, and in the copy number variation dataset, i.e. genes which exhibited copy number amplification and gene expression upregulation, or genes which exhibited copy number deletion and gene expression downregulation. Using this recipe we have identified 48 genes matching this description, which are the stemness regulators.
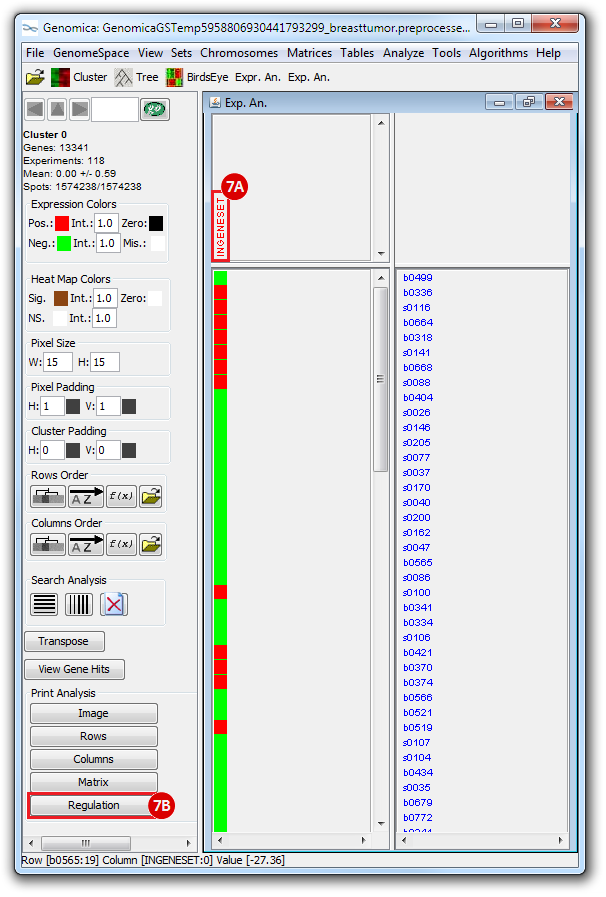
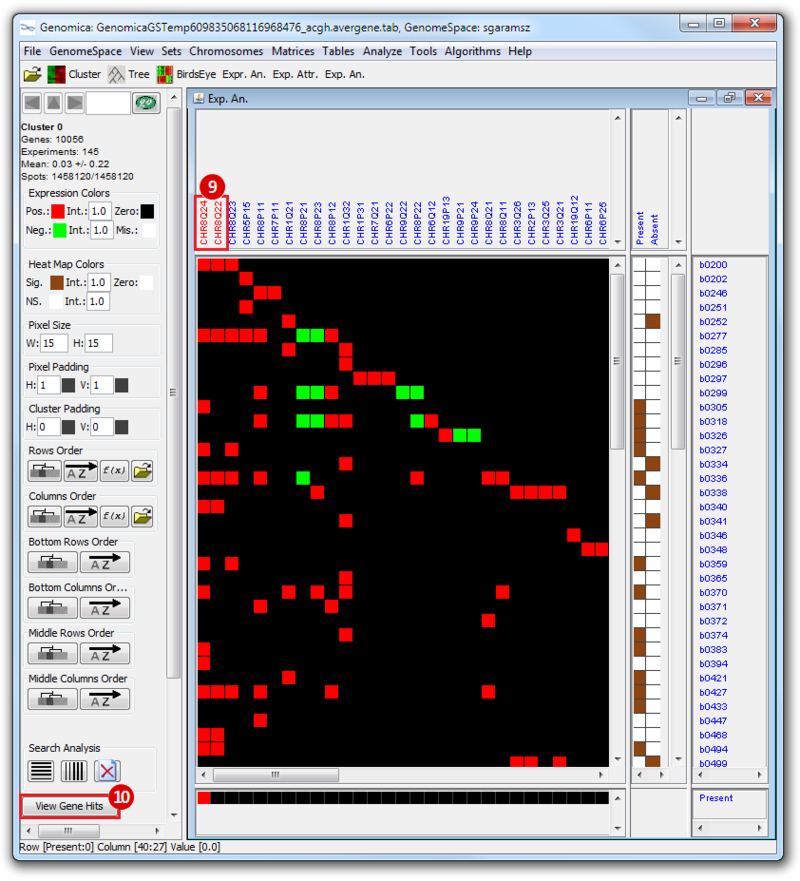
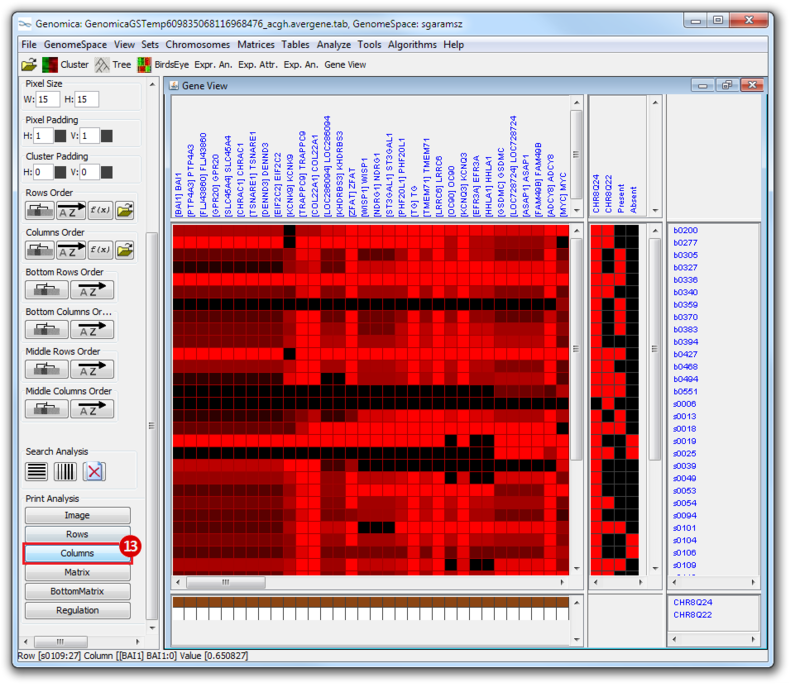
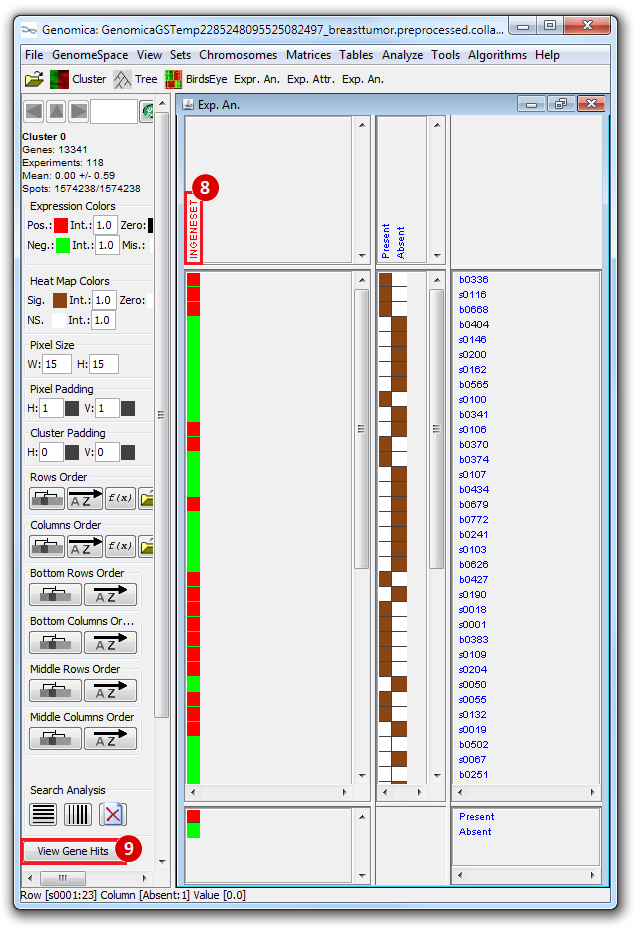
The heatmap below illustrates the expression of the stemness regulators in breast cancer tumor samples. Red indicates upregulation of expression, green indicates downregulation of expression. Gene names are listed to the right of the heatmap, and the sames of breast cancer tumor samples are listed above the heatmap. Note that only breast cancer tumor samples with the presence or absence of the stemness signature are included. To the left, a single column indicates which genes are in the ‘INGENESET’ category, which is the label for regulators of the copy number variation data. In this example, all genes fall into that category, i.e. they are associated with both copy number variation, and gene expression. The rows on the bottom of the heatmap indicates which breast cancer tumor sample falls into which category. In the first row, red indicates ‘presence’, and green indicates ‘absence’, in the second and third rows, red indicates ‘presence’. These are not related to gene expression changes.
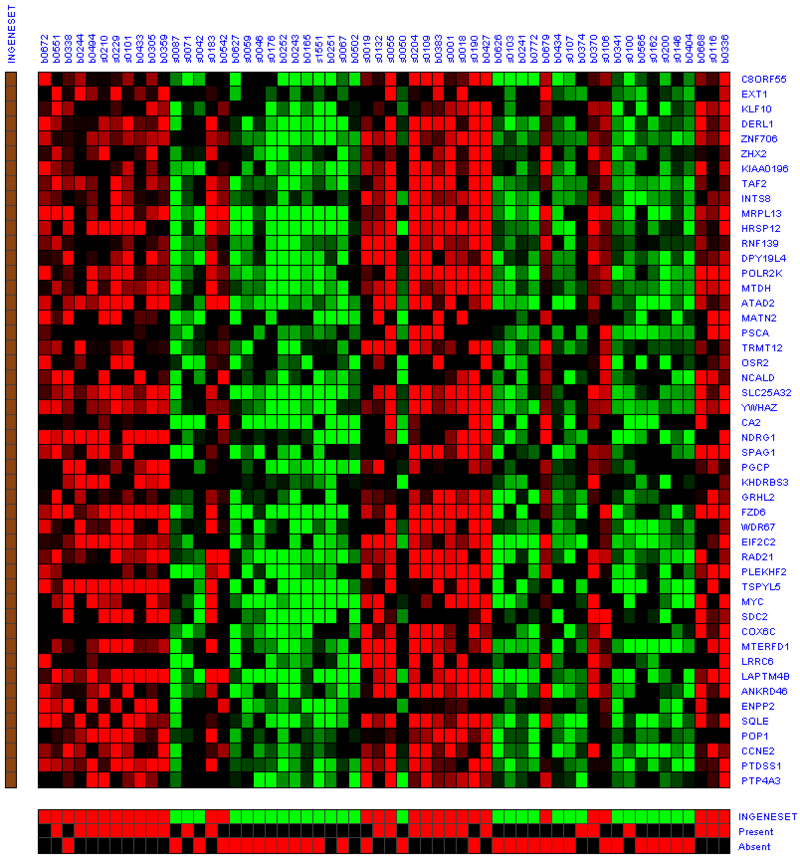
Notice that these stemness regulatory genes tend to have the same overall expression patterns within one sample, i.e., both TAF2 and MYC are downregulated in sample b0404 (right-most green sample). Note that the pattern exhibits concordance between the stemness signature, and the stemness regulator. Sample b0404 lacks the stemness signature, and appears to have downregulation of stemness regulatory genes. In contrast, sample b0668 has the stemness signature present, and appears to have upregulation of stemness regulatory genes.
- To save the list of stemness regulators from Genomica:
- Under
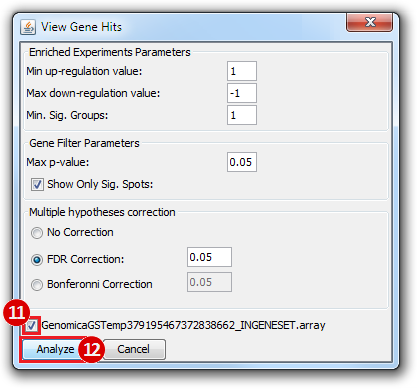
Print Analysis, click Columns.
- Save the resulting file of 48 stemness regulators as, e.g.,
stemness_regulators.lst.
- Click
Save.
- You can save this file to GenomeSpace by clicking and dragging the files into your GenomeSpace directory from your local directory.
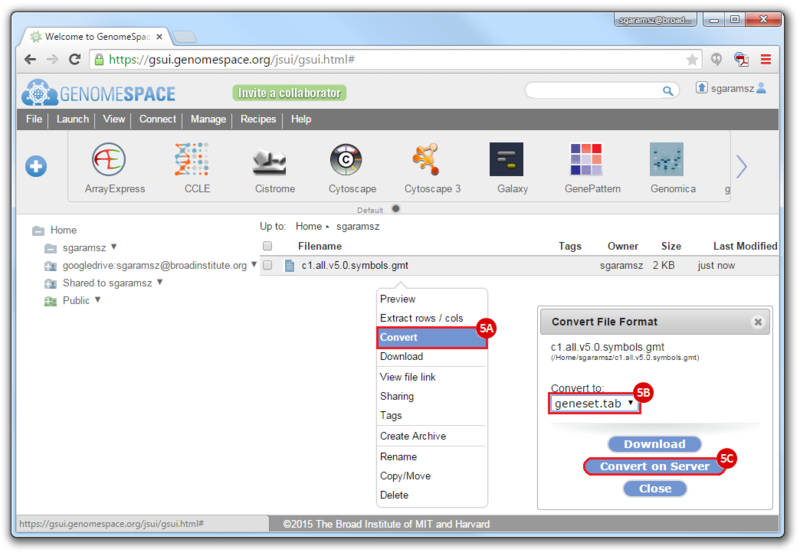
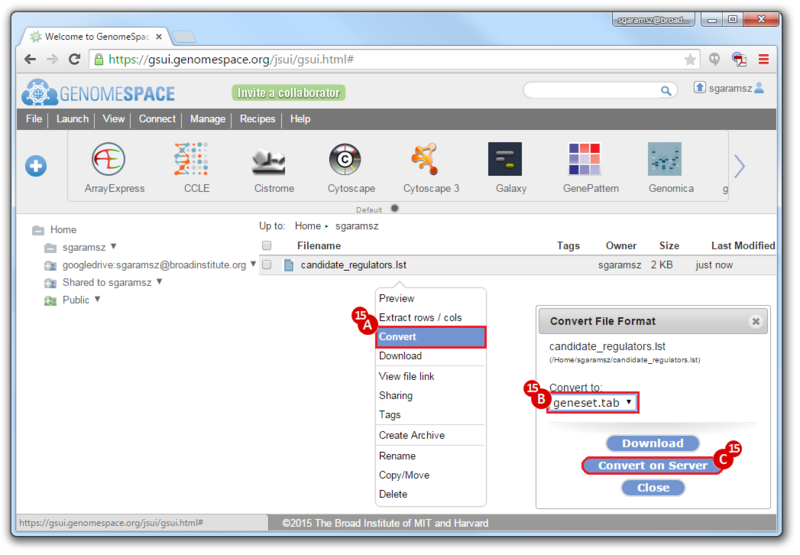
- To convert the LST format to a Genomica geneset TAB format:
- Right-click on the file
- Choose
Convert
Convert to: geneset.tab- Click
Convert on Server
 GenomeSpace.org
GenomeSpace.org